If you’ve ever wished there was GO but for phenotypes instead of gene functions, check out uPheno!
In a preprint just made available in bioRxiv, Matentzoglu and many past/present GO Consortium members and Alliance representatives detail their work on the new Unified Phenotype Ontology (uPheno).
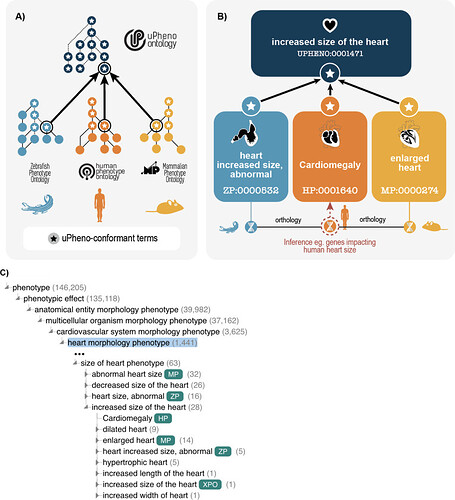
uPheno is a framework for consistent and logical definition of phenotype categories. uPheno currently integrates 12 species-specific phenotype ontologies and helps make datasets interoperable across species:
The uPheno ontology is a logic (OWL) based ontology that combines existing phenotype ontologies into a single ontology and introduces common grouping classes such as UPHENO:0082544 “mitochondrion phenotype”. For example, HP:0001640 “Cardiomegaly”, MP:0000274 “enlarged heart” and ZP:0000532 “heart increased size, abnormal” all classify under a common species-neutral grouping UPHENO:0001471 “increased size of the heart”, which is in turn classified under UPHENO:0075162 “size of heart phenotype”.
uPheno is used by or will soon be used by the International Mouse Phenotyping Consortium (IMPC), MGI, and the Monarch Initiative, and other influential groups.
![]() Explore uPheno using OLS: Ontology Lookup Service (OLS)
Explore uPheno using OLS: Ontology Lookup Service (OLS)
![]() Find the bioRxiv paper here: The Unified Phenotype Ontology (uPheno): A framework for cross-species integrative phenomics
Find the bioRxiv paper here: The Unified Phenotype Ontology (uPheno): A framework for cross-species integrative phenomics