In a new PLOS Biology publication, Baijal et al. investigate the role of polyphosphate kinase (PPK) and the pathways regulated by PPK in E. coli.
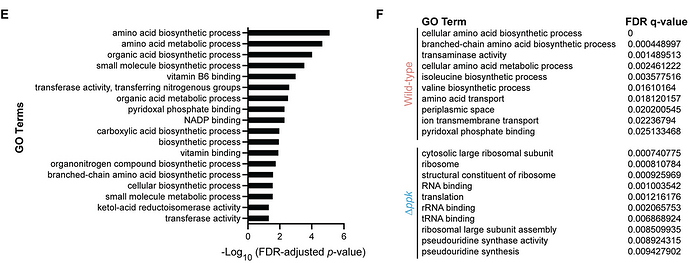
We performed Gene Ontology (GO) [30] enrichment analysis on the significantly differentially expressed and all-or-none proteins (92 total) and identified 16 enriched GO terms (FDR-adjusted p-value < 0.05) (Fig 1E). These included terms related to amino acid, organic acid, and small molecule biosynthesis. We next used Gene Set Enrichment Analysis (GSEA) [31] on the entire data set of 1,909 proteins to look for GO terms that are differentially expressed between the wild-type cells and Δppk mutants. This analysis showed that wild-type cells were enriched for proteins related to amino acid biosynthesis and transport (Fig 1F). In contrast, Δppk mutant cells were enriched for proteins broadly related to ribosome biogenesis and translation (Fig 1F)… [T]hese data point to a model wherein Δppk mutants fail to properly respond to starvation by remaining primed for growth while failing to activate pathways to increase the availability of amino acids and other biomolecules needed for that purpose.
Not only will this paper be of extreme interest to anyone studying how bacterial cells respond to starvation, it also earns a gigantic ![]() for completely citing GO. Baijal et al. include a GO reference publication, the tool used, the release (2023-02-13) for ontology and annotation files, and describes the background used for EA- ensuring the work is reproducible.
for completely citing GO. Baijal et al. include a GO reference publication, the tool used, the release (2023-02-13) for ontology and annotation files, and describes the background used for EA- ensuring the work is reproducible.