The mission of the GO Consortium is to develop a comprehensive, computational model of biological systems, ranging from the molecular to the organism level, across the multiplicity of species in the tree of life.
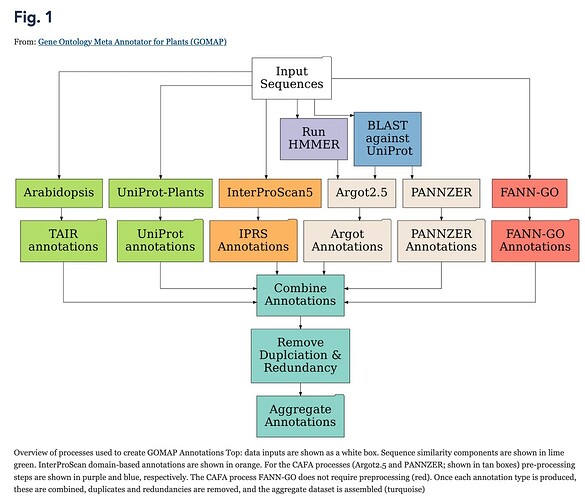
Although many researchers on the Alliance of Genome Resources pages are looking for information on major model organisms, that’s not even near the limit of GO. In a recent BMC Research Note publication, Fattel et al. have annotated the latest published sequences of 26 maize lines using GOMAP: Gene Ontology Meta Annotator for Plants. This tool is yet another innovative tool developed by an outside-GO group and in the GO-MAP paper in Plant Methods, Wimalanathan & Lawrence-Dill note that “GOMAP expands and improves the number of genes annotated and annotations assigned per gene as well as the quality (based on ) of GO assignments in maize. GOMAP has been deployed to annotate other species including wheat, rice, barley, cotton, and soy”. GO has not examined this tool but it looks incredibly promising!
Find the 26 Zea mays Nested Association Mapping (NAM) founder line annotation paper here.