Homologous recombination is a universal pathway to repair DNA double-stranded breaks, ssDNA gaps, and stalled or collapsed replication forks. Homologous recombination relies on an intact dsDNA donor that is identical to the broken molecule to template the repair event in a copy/paste-type reaction. Typically, this donor is the allelic region on either the sister chromatid or homolog. But what happens when DNA damage falls within, or proximal to, repetitive DNA? A study recently published in Genes and Development by Diedre Reitz and colleagues shows that, in rare cases, homologous recombination can engage two or more repetitive elements in a process termed multi-invasion recombination.
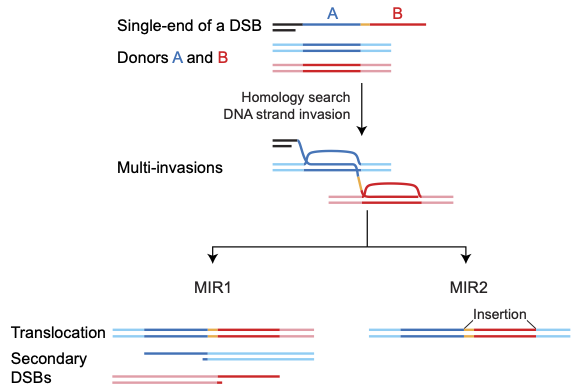
Model of multi-invasion-induced rearrangements. MIR1 generates a translocation and two additional single-ended DSBs. MIR2 generates an insertion.
Using genome-wide sequencing, Reitz and colleagues found that multi-invasion recombination can lead to a cascade of recombination-induced rearrangements, aneuploidies, and secondary DNA breaks. By creating a specialized reporter assay, they discovered two pathways in which multi-invasion intermediates can be resolved into products, thereby forming multi-invasion-induced rearrangements. The MIR1 pathway can occur in any sequence context, generates secondary DSBs, and frequently leads to additional genome rearrangements. The MIR2 pathway occurs only when the recombining donors exhibit substantial homology, and results in an insertion without additional DNA breaks. To better understand the molecular determinants of MIR1, the authors developed a highly sensitive proximity ligation-based assay to detect rare MIR1 translocation events. Using this assay, the authors found that, in contrast to normal repair, MIR1 does not require displacement DNA synthesis to fill in the nucleotides lost due to DNA damage.
These vast rearrangements originating from a single DNA break and resolution via the MIR1 pathway are reminiscent of chromothripsis, a mutational signature characterized by large clustered genomic rearrangements frequently associated with certain cancers. Overall, the findings by Reitz and colleagues provide new mechanistic insight into how complex recombination-dependent rearrangements can occur.
– Text from Diedre F. Reitz, with edits and link outs from SGD.