Eroglu, Yu and Derry (1) showed convincingly that long ssDNAs are great repair templates for doing CRISPR edits in worms.
Obtaining good yields of ssDNA is important for this strategy.
I read in the NEB catalog that phosphorthioate linkages are resistant to T7 5’->3’ Exonuclease.
The recommendation is to put PT linkages between the first 5 bases at the 5’ end of one of your PCR primers (full-disclosure, I did 6). PT modified oligos are easy and relatively cheap to order from IDT and other suppliers (indicate by asterisks between the bases when you input the sequence, e.g. G*C).
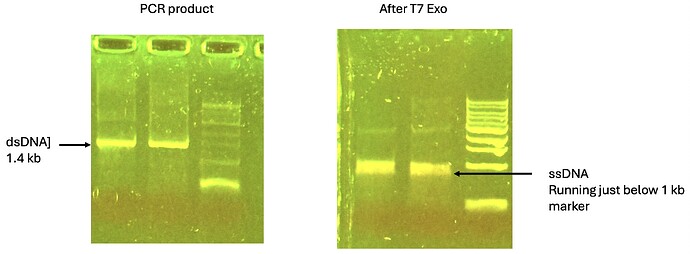
I column purified my PCR product before doing the digestion, but purification might not be necessary. I did the reaction in NEB4 (about 5 micrograms in a 30 ul reaction with 1 ul of NEB T7 Exo and left it overnight at 25 C. NEB recommends 25 C, but their tech support person said 37 is also okay (should be faster). In my trial experiment, essentially all of the ds DNA was converted ssDNA.
Searching online, I found that this had already been worked out a few years ago, so I recommend looking at Noteborn et al. (2) for a comparison of different ssDNA prep methods.
- Eroglu M, Yu B, Derry WB. Efficient CRISPR/Cas9 mediated large insertions using long single-stranded oligonucleotide donors in C. elegans. FEBS J [Internet]. 2023 May 31; Available from: http://dx.doi.org/10.1111/febs.16876
- Noteborn WEM, Abendstein L, Sharp TH. One-pot synthesis of defined-length ssDNA for multiscaffold DNA origami. Bioconjug Chem. 2021 Jan 20;32(1):94–8.