Hello all,
I am facing issues with the SWPCR of the heterozygous strain, which is made by crossing a single mutant (deletion of 1kb) strain with another homozygous strain (superficially wt but having m-cherry by MosSCI on the same chromosome as that of mutation). I expect the F1 progeny to have two bands i.e. for deletion mutation and wt, but on performing PCR, I get 4 bands, out of which two are as expected, but the other two bands are always there.
I have checked the specificity for primers, and they seem to work fine. Anyway, the F2 progeny for the same crossed strain shows only a single band for the desired mutation. Please suggest if anyone has faced this problem.
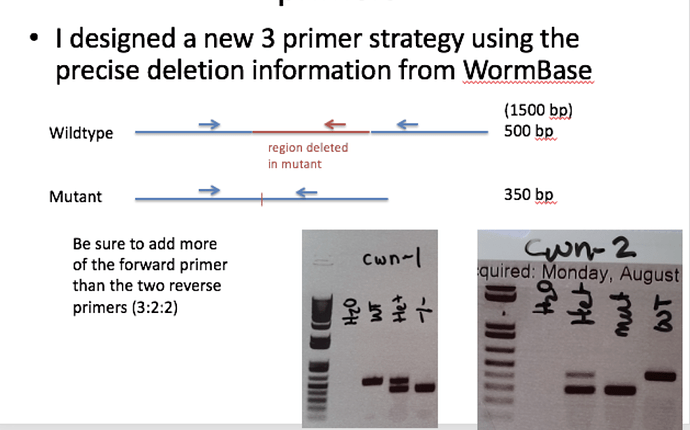
We use a 3 primer strategy for deletion alleles. Below is the intro for the instructions on how to design the primers. Please let me know if you need more detailed instructions.
Deletion alleles created by the Knockout consortium often have primer information included in the description on Wormbase. These primers amplify long regions because they were used to detect deletions that occurred. They are not good for genotyping–if the deletion is more than a few hundred bp, the wildtype product is much longer and hard to detect in heterozygotes because it is thermodynamically unfavorable compared to the shorter mutant product. I highly recommend you design your own primers using a three primer strategy to mitigate the size difference between the WT and deletion alleles. Basically you design two sets of primers with the same forward primer, one reverse primer on the opposite side of the deletion and one reverse primer that sits within the deletion. For the wildtype allele, the closer reverse primer will produce a product that is more thermodynamically favorable than the far reverse primer on the other side of the deletion region and you will hardly see the longest band, if at all. **Since in most cases, you are trying to identify a homozygous mutant, it’s better to make the WT band shorter than the mutant (opposite of the examples above) to be more sure that it will show up and you won’t accidentally select a het for further study.
**
Another idea: we “simulate” hets for SWPCR by picking a WT and a homozygous mutant into the same tube for SWPCR. Maybe you could try that so you have “heterozygote” DNA to optimize your PCR conditions with.