Our Gene Ontology overview page goes over how the GO is a loosely hierarchical graph, with GO terms as nodes and the relationships between terms are the edges between nodes. In the GO, ‘child’ terms are more specialized than their ‘parent’ terms, but unlike a strict hierarchy, a term may have more than one parent term (note that the parent/child model does not hold true for all types of relations).
Understanding the ontology is critical to understand the meaning of an annotation. Most annotations propagate up the ontology.
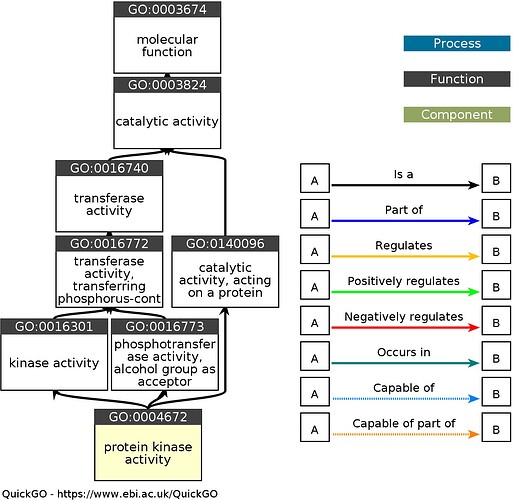
In the below Ancestor Chart (available for all GO terms at QuickGO), it is clear that GO:004672 protein kinase activity has several is_a ancestors. Therefore, for any gene product with an annotation to GO:004672 protein kinase activity, an equally true statement could be made by an annotation to GO:0016301 kinase activity or any other is_a ancestor.
Absence of an annotation does not imply absence of a function, so no information about child terms of GO:004672 protein kinase activity would be conveyed by this annotation.
However, there is an exception to annotations propagating up -any annotation that uses the NOT modifier. The NOT modifier was previously mentioned in another post. Contrary to positive annotations, NOT statements propagate down the ontology, such that the annotation gene product NOT|enables kinase activity means that the gene product does not enable GO:004672 protein kinase activity or other child terms either.
Full guidelines for NOT are on the GO wiki.