GO’s PAN-GO Functionome, just announced last week (ICYMI: GO's new human gene resource and accompanying Nature paper on the "PAN-GO Functionome" is here!), can be used in many different ways. This how-to is about finding the functions (including cellular location and broader processes) of a specific human gene.
Only have a minute? Here’s how to use the PAN-GO Functionome to view gene functions, in less than 60 seconds!
![]() Remember, if you have any questions, comments, or change requests to data in the PAN-GO Functionome, your feedback is valued: GO Functionome Feedback Form.
Remember, if you have any questions, comments, or change requests to data in the PAN-GO Functionome, your feedback is valued: GO Functionome Feedback Form.
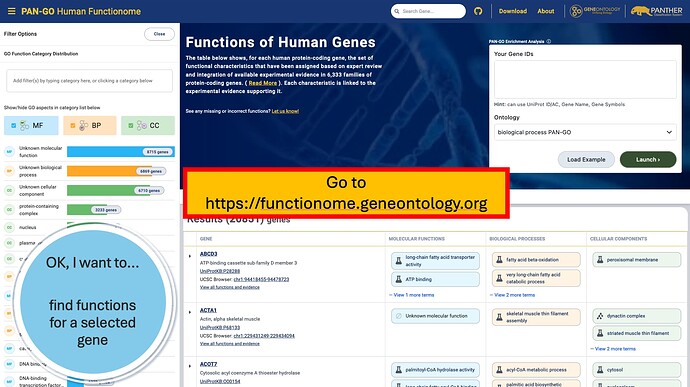
Detailed guide on finding functions for a human gene:
Searching the PAN-GO Functionome
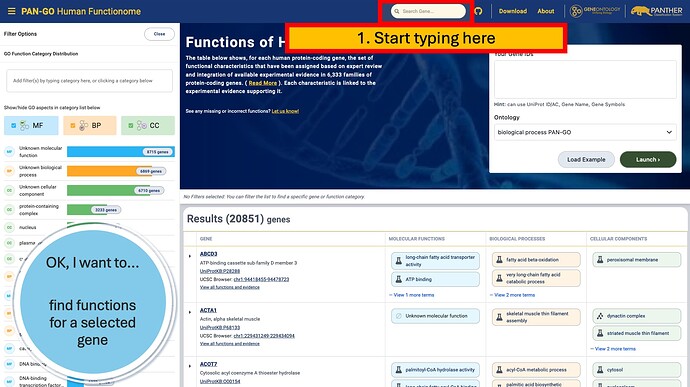
The PAN-GO Functionome is at https://functionome.geneontology.org/
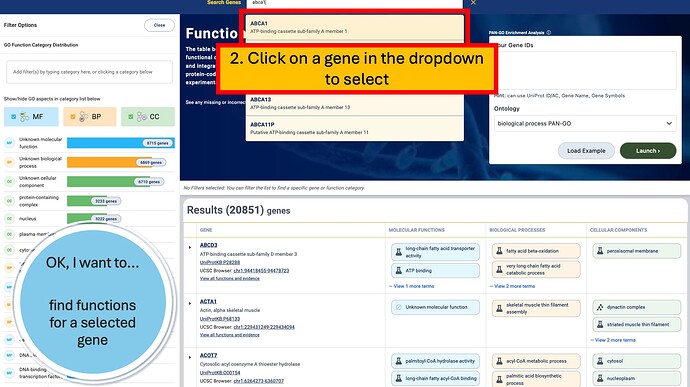
You can search for gene names, protein names, UniProt identifiers (e.g. Q9NWH9 ), and more.
The search field will autosuggest genes to help you locate the correct entry.
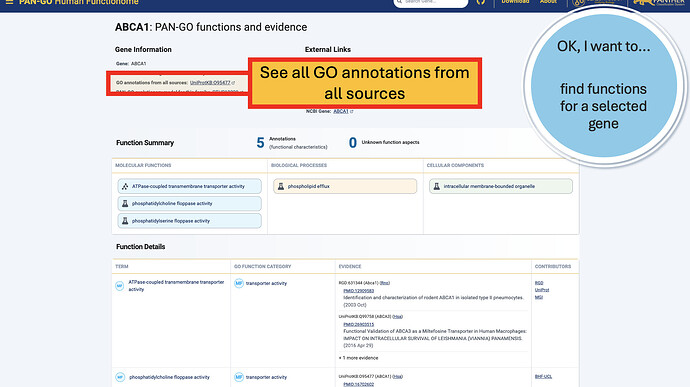
PAN-GO Functionome Gene Pages
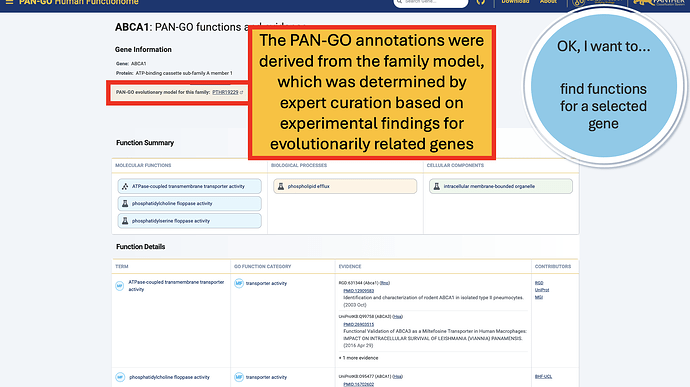
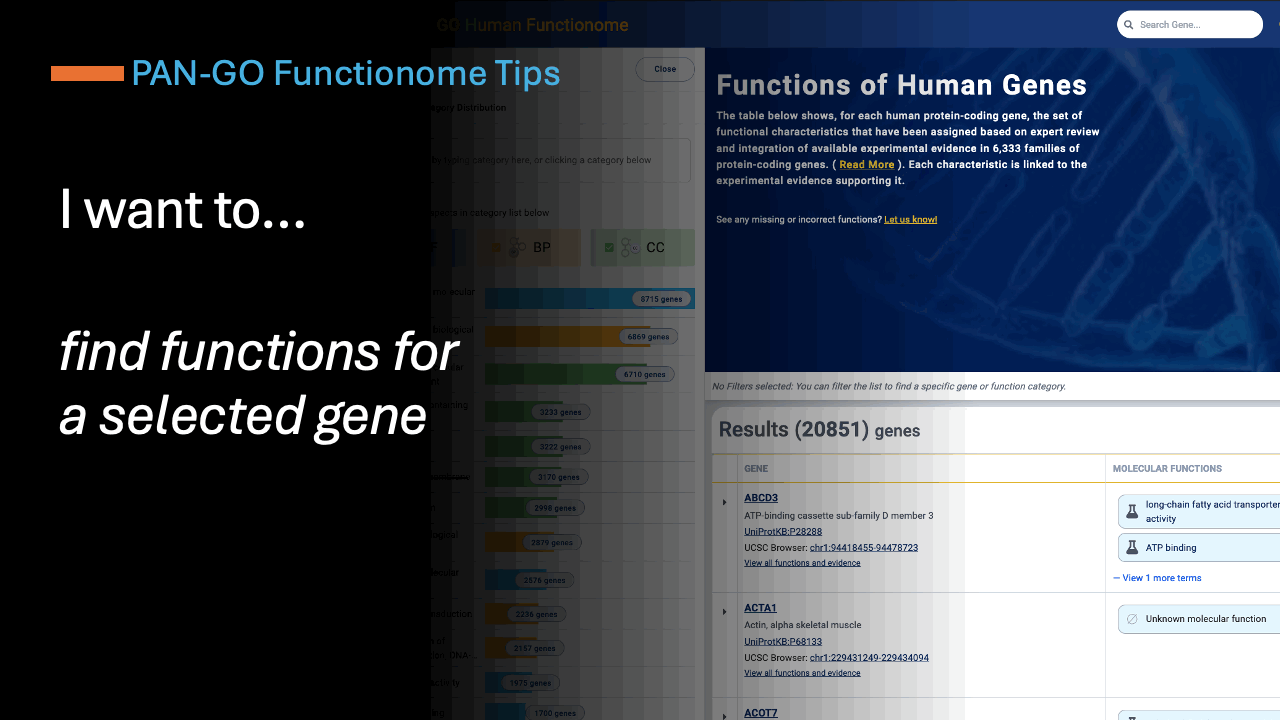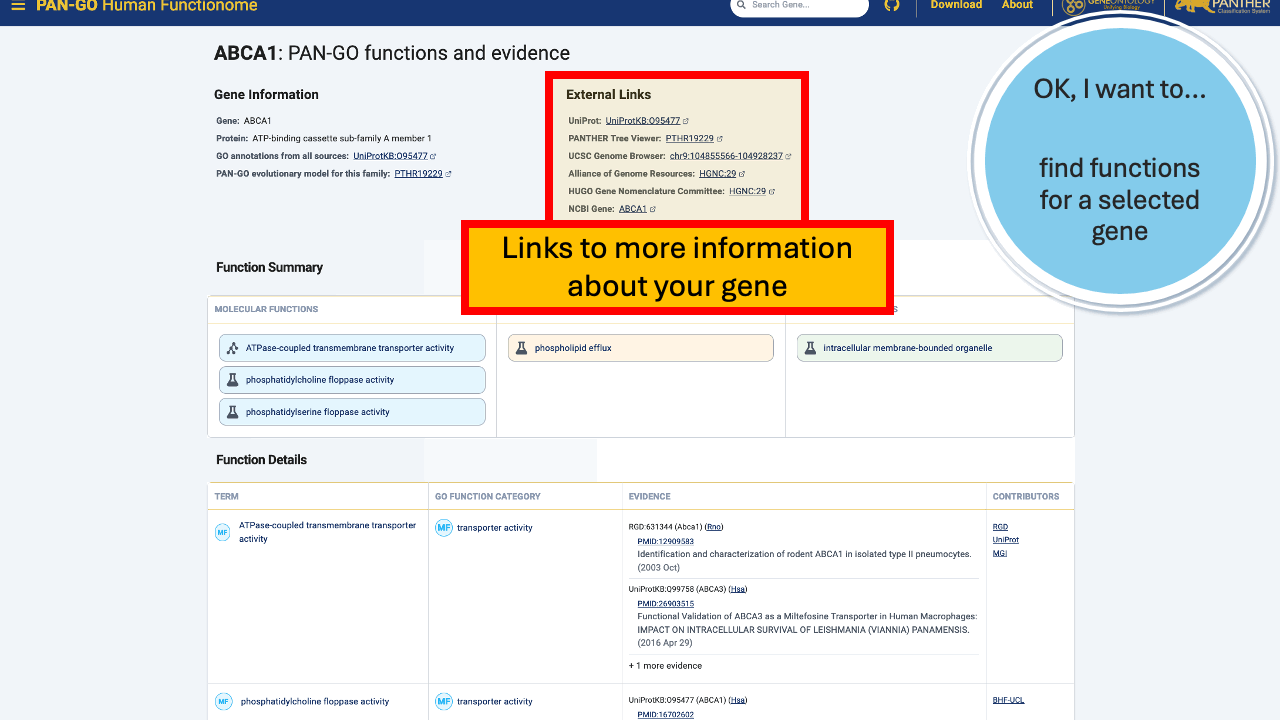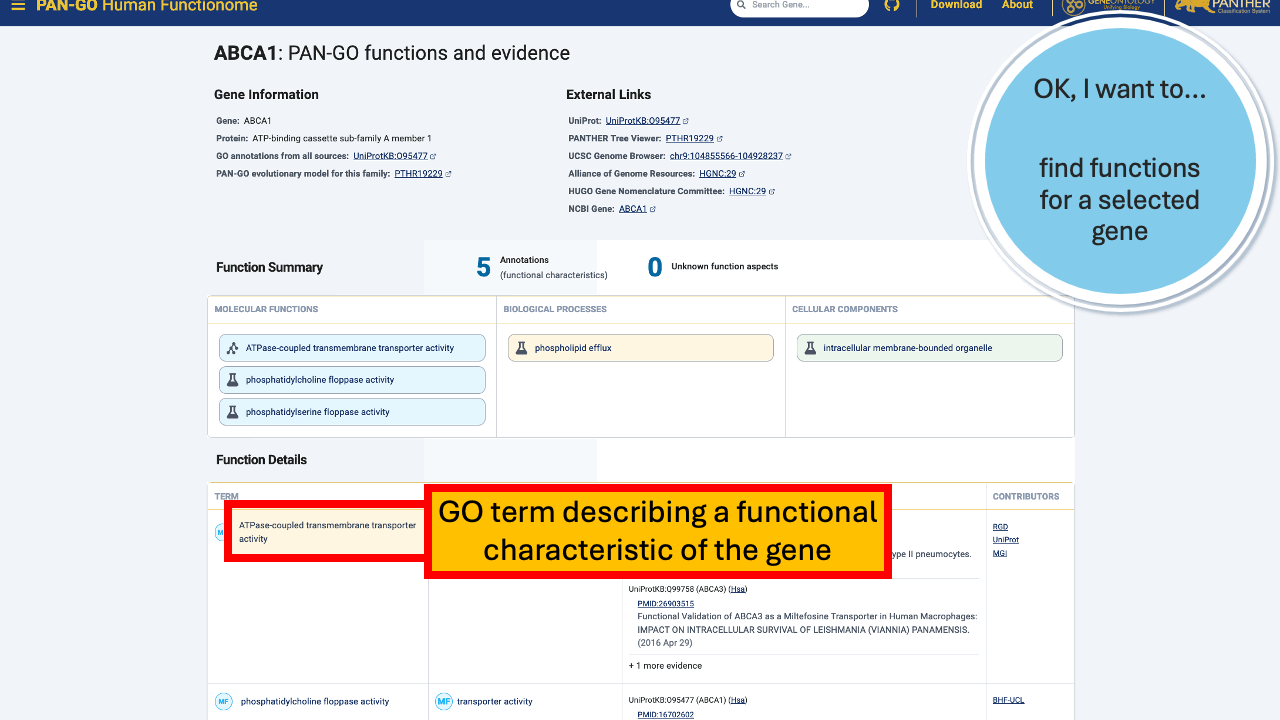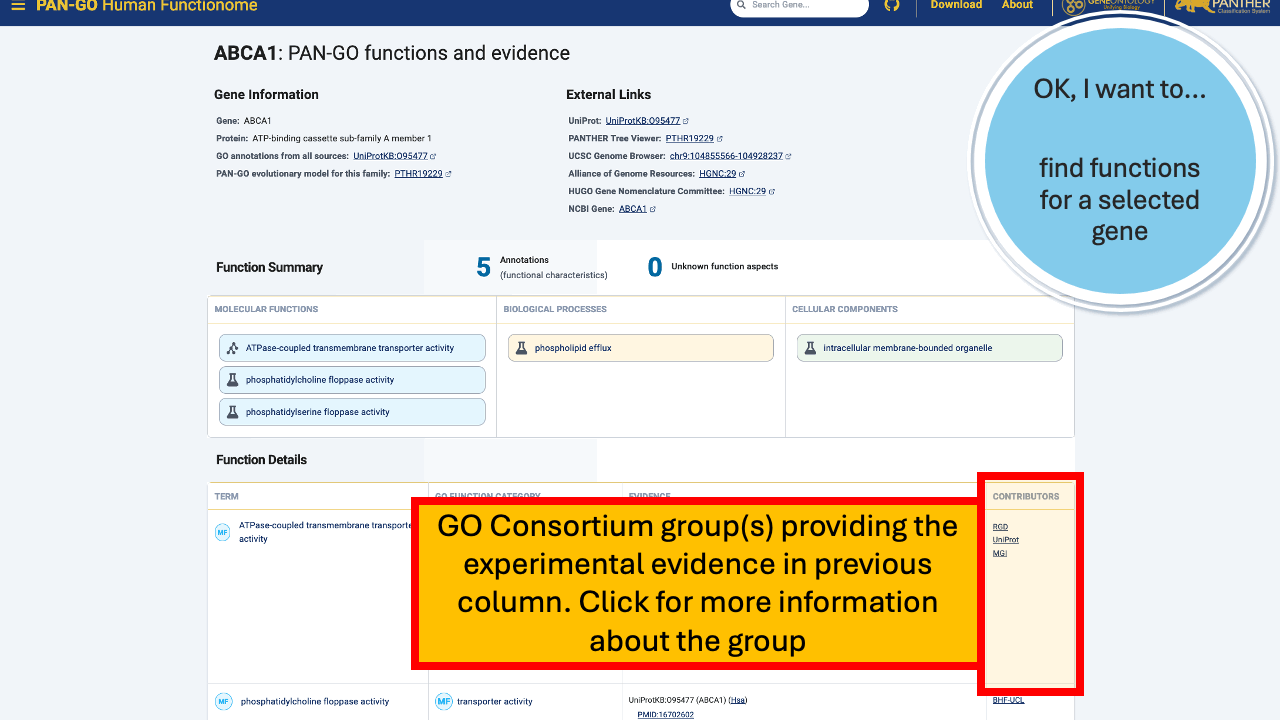
Once you’re on a gene page, you’ll notice some different sections. The top Gene Information section includes gene and protein names, a link to all annotations, and a link to the PAN-GO model that the annotations are based on.
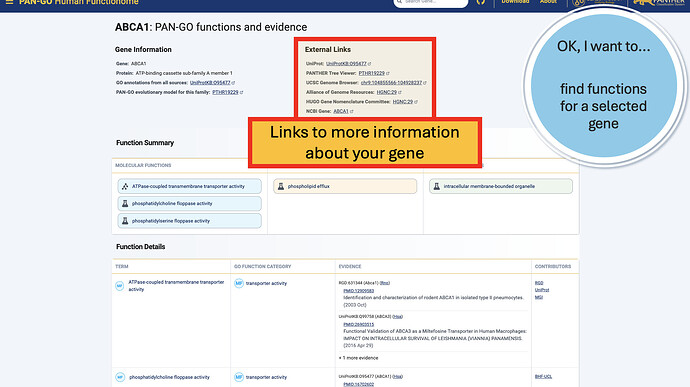
Additionally, the External Links gives you quick access to other knowledgebases and browsers, including Alliance links, the UCSC Genome Browser, NCBI gene page, and more.
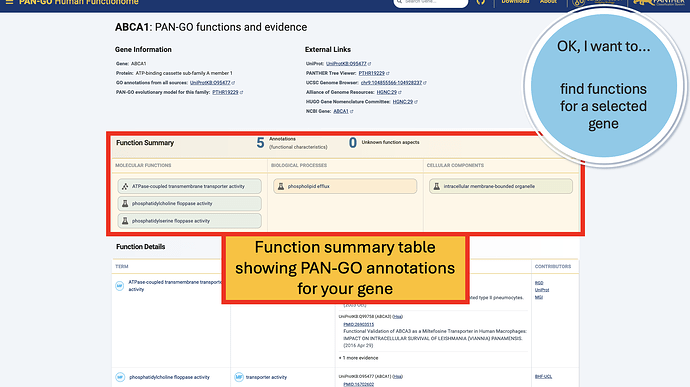
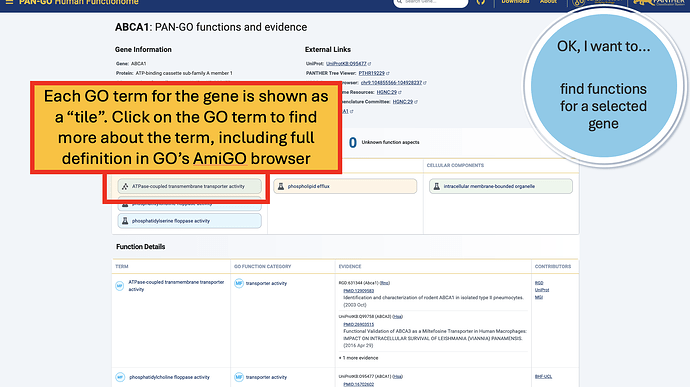
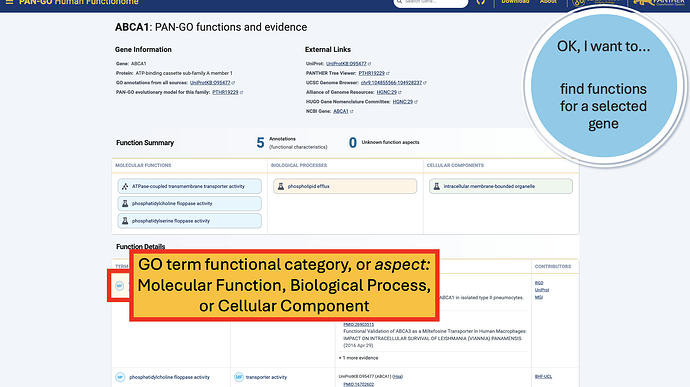
The Function Summary has tiles divided by Molecular Function, Biological Process, and Cellular Components. These are the three Function Categories, also referred to as “aspects”. The icon on the left of each icon indicates the type of evidence: a flask indicates direct evidence, and a tree icon indicates homology-based evidence. If there’s no known function, you’ll see a universal no (Ø) and a note that it’s “unknown” molecualr function, biolocal process, or cellular component. An unknown can occur in only one, only two, or all three categories.
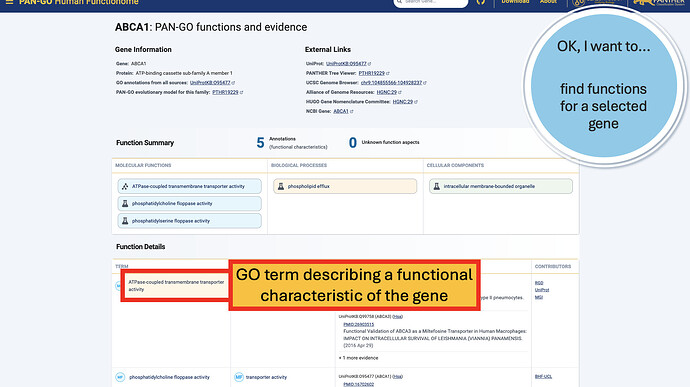
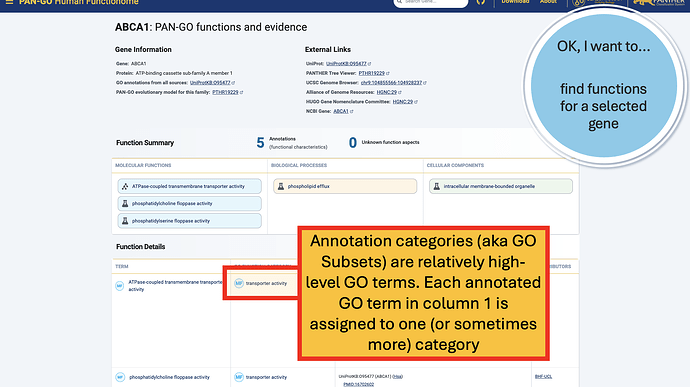
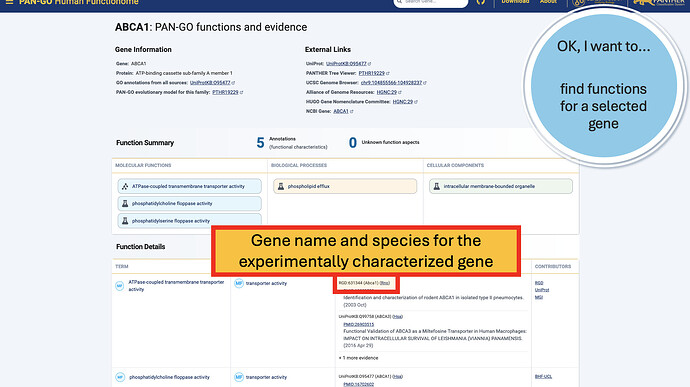
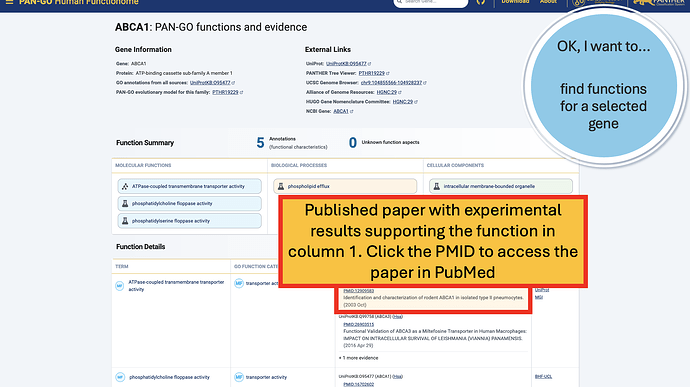
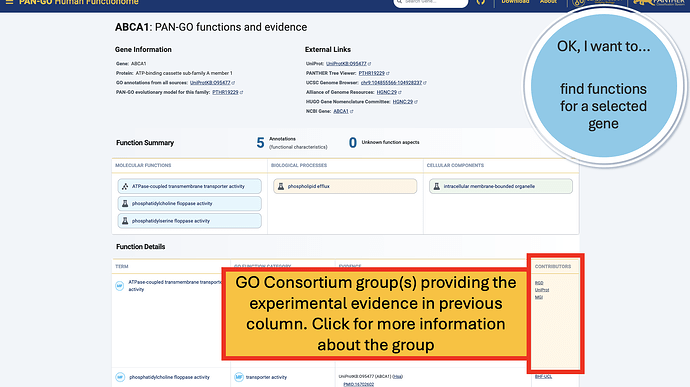
The Function Details section provides more information on each annotation. You can find the specific GO term, as well as a Annotation Category term that summarizes the GO term and can be used to make general lists- experienced GO users have seen these before as GO Subsets or GO Slim terms. The Evidence column contains links to the exact publication annotations are based on, as well as details about the organism, gene, and database that provided the experimental annotation the PAN-GO annotation is based on.
Again, if you have any questions, comments, or change requests to data in the PAN-GO Functionome, your feedback is valued: GO Functionome Feedback Form.