Is your reference sequence in NCBI?
If your research is on an organism that’s not well annotated but has a reference sequence in NCBI, there’s a simple way to retrieve GO annotations: NCBI’s RefSeq FTP.
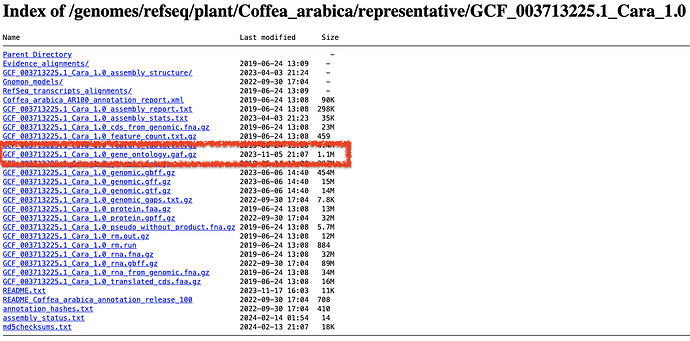
Start at https://ftp.ncbi.nlm.nih.gov/genomes/refseq/. Navigate to your organism, then find the genome in the /representative/ directory:
From there, you can download the file with the suffix gene_ontology.gaf.gz. You’ll now have a GAF 2.2 that contains electronically inferred annotations (IEA) assigned by RefSeq!
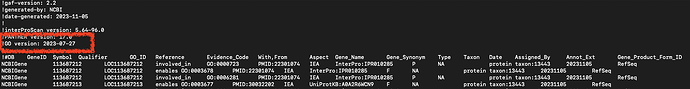
Now, if you need the corresponding ontology file, check the header to get the GO version used in the construction of the GAF. Here, it’s !GO version: 2023-07-27, which like all other releases can be found in the GO Release archive.
Read the full NCBI announcement to learn more about this pipeline that uses InterProScan and TreeGrafter at PANTHER.
Annotations for well annotated/model organisms
If you’re investigating an Alliance organism (fly, mouse, rat, budding yeast, worm, frog, or zebrafish), finding the GO annotation file is fairly trivial:
- pop on over to the official GO Downloads page and grab the GAF for your organism of interest
- GO have a coffee, because that’s it!
Unannotated genomes
See the previous post about generating brand new annotations for an unannotated organism.